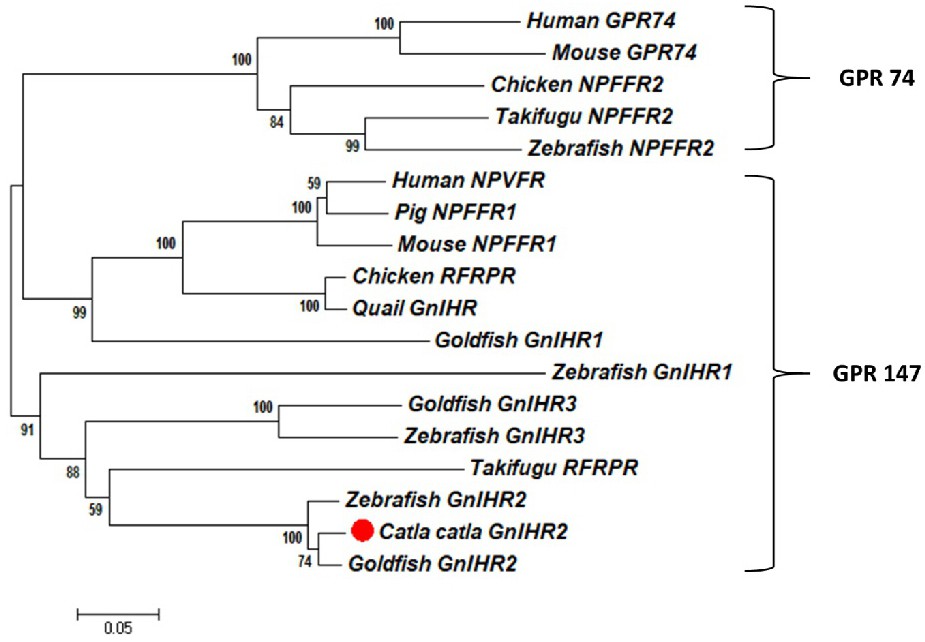
Fig. 4. Phylogenetic relationship of deduced amino acid sequences of Labeo catla GnIHR2 with vertebrate species was conducted in MEGA7 software. The consensus tree was inferred using the Neighbour-Joining algorithm, and the branch points were validated by 1000 bootstrap replications. The position of catla GnIHR2 was marked with a solid circle sign. The GenBank accession numbers of sequences for analysis are as follows: human (Homo sapiens) GPR74 (AAK58513), mouse (Mus musculus) GPR74 (AAK58514), chicken (Gallus gallus) NPFFR2 (NP_001029997.2), Takifugu rubripes NPFFR2 (NP_001092118.1), zebrafish (Danio rerio) NPFFR2 (XP_690069.6), human (Homo sapiens) NPVPR (AAK94199.1), pig (Sus scrofa) NPFFR1 (NP_001193386.1), mouse (Mus musculus) NPFFR1 (NP_001170982.1), chicken (Gallus gallus) NPFFR (BAE17050.1), quail (Coturnix japonica) GnIHR (BAD86818.1), Takifugu rubripes NPFFR (NP_001092117.1), goldfish (Carassius auratus) GnIHR1,2 and 3 (AFY63167.1, AFY63168.1 and AFY63169.1), zebrafish (Danio rerio) GnIHR1,2 and 3 (ADB43133.1, ADB43134.1and ADB43135.1).